import math
import matplotlib.pyplot as plt
import numpy as np
import random
import seaborn as sns
import pandas as pd
!pip install lmfit
from lmfit import Model
import os.path
import subprocess
from numpy.fft import fft, ifft
def wget_data(url):
local_path='./tmp_data'
subprocess.run(["wget", "-nc", "-P", local_path, url])
Fourier Methods#
Fourier Methods involve two pieces:
Fourier Series
Fourier Transforms
Both are extremely powerful for finding patterns in data. This is an extremely broad topic.
Fourier Series
We can write a Fourier series as:
for a periodic function \(f(x)\) which has a finite number of finite discontinuities and only a finite number of extrema over the range of 0 to \(2\pi\). The coefficients come from the integrals: $\(a_n = \frac{1}{\pi} ∫_0^{2\pi}f(t)\cos nt dt\)\( and \)\(b_n = \frac{1}{\pi} ∫_0^{2\pi}f(t)\sin nt dt\)\( Then the sums will form a complete orthoganal set and \)f(x)$ can be specified with any desired accuracy by knowing the coefficients here.
There are many such periodic functions in physics and so this has wide applicability.
Here we’re going to focus on extracting these correlations from data. I’m going to use the specific example of heavy-ion collisions (my field) but the idea of extracting information from a limited, fluctuating dataset is not unique.
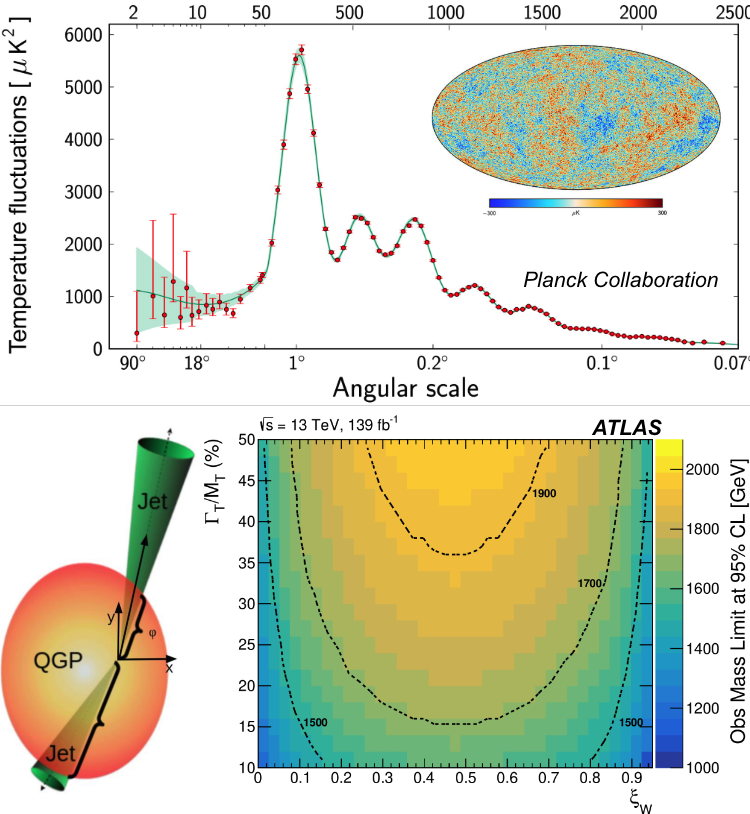
Physics Motivation In relativistic collisions between two nuclei, the coefficents Fourier coefficients, \(v_n\), are proportional to the eccentricities of the initial state of the overlap region between two nuclei. Here is a simulation of such a collision projected onto the x-y plane.
The constant of proportionality is related to the viscosity of the matter created in these collisions (exact details due to relativistic viscous hydrodynamics). Measuring these \(v_n\) values, is the most direct experimental method to constrain the viscosity of the matter created in heavy-ion collisions.
You can think of expanding the shape of the overlap region in terms of \(n-\)th order eccentricities.
In reality, determination of the Fourier coefficients can be unstable. Here is an example of how that can be…
Let’s say we’ve got \(N\) particles in an event. We can clearly describe the particle distribution by a Fourier series: $\(\frac{dN}{d\phi} ∝ 1 + \sum_{n=1}^{∞} 2v_n\cos(n(\phi-\Psi_n))\)\( Instead of a \)\sin\( and \)\cos\( term we now have \)n\( phases, \)\Psi_n\( and just the \)\cos$ term.
However, we don’t know what \(\Psi_N\) is so we have to determine it from the data itself. Determining \(v_n\) directly as above is difficult mainly because each event is differet from any other event and each single event has a finite number of particles.
Here is a plot from real data of \(\frac{dN}{d\phi}\) paper. Thre specific events are shown; each event has about a 1000 particles (error bars are the \(\sqrt{N}\) errors we have discussed). Impossible to constrain \(v_n\) and the \(\Psi_n\) values in these events.
Determining Coefficients from Data
A more direct way to deal with this issue is to use correlations. If we can describe the angular distribution of single particles as: $\(\frac{dN}{d\phi} ∝ 1 + \sum_{n=1}^{∞} 2v_n\cos(n(\phi-\Psi_n))\)\( then we can describe the distribution of pairs of particles as: \)\(\frac{dN^{ij}}{d\phi_i d\phi_j} ∝ [1 + \sum_{n=1}^{∞} 2v_n\cos(n(\phi_i-\Psi_n))][1 + \sum_{n=1}^{∞} 2v_n\cos(n(\phi_j-\Psi_n))]\)\( \)\(\frac{dN^{ij}}{d\phi_i d\phi_j} ∝ 1 + \sum_{n=1}^{\infty} 2v_n^i v_n^j \cos(n(\phi_i - \phi_j))\)\( Now this is in terms of things we can directly measure, the angles of individual particles. The rhs only depends on the difference between the angles so we can define \)\Delta \phi \equiv \phi_i - \phi_j\( and rewrite the equation as: \)\(\frac{dN^{ij}}{d\Delta\phi} ∝ 1 + \sum_{n=1}^{\infty} 2v_n^i v_n^j \cos(n(\Delta\phi))\)$
Here are the same three events from above, now showing the \(\frac{dN}{d\Delta\phi}\) distribution as well. This looks clearer (1000 data points yield about 1000000 pairs).
So we’ve eliminated a hard to determine phase by only looking at the angular difference between pairs of particles. Constructing a double loop over each pair of non-identical particles, we can construct \(\frac{dN^{ij}}{d\Delta \phi}\). This is a correlation function. This is well suited to constraining the coefficients of the Fourier series (as we’ll see below) but correlations functions are used in many scenarios (for a more formal districription see Pruneau Data Analysis Techniques for Physical Scientists.
For our purposes we don’t even care about the normalization of this correlation function. Exactly this quantity was measured in lead-lead collisions at the LHC and the first eight terms of the Fourier series were extracted (paper). Here \(V_{n\Delta} = v_n^i v_n^j\) from our notation.
This is amazing; under the assumption that \(v_n \propto \varepsilon_n\) then we are really measuring the shape of the nuclear overlap and we are decomposing it in terms of the various ordered harmonincs.
However, when two-particle correlations are measured, what they are actually sensitive to is \(v_n^iv_n^j\). This causes some complications in extracting \(v_n\).
First, let’s take a look at some data generated with:
100 events
50 particles in each event
every particle in event having exactly the same \(v_2\) value (\(v_2 = 0.25\))
all other \(v_n\) values are zero
Here is some (inefficient) code to pair every particle with every other particle from the same event. We only pair particles from the same event because those are the ones that the math above allowed for.
wget_data('https://courses.physics.illinois.edu/phys398dap/fa2023/data/vn_sigma00.txt')
data = pd.read_csv('tmp_data/vn_sigma00.txt', header=None)
print(data)
import itertools
dphi_vals = np.array([])
for row1, row2 in itertools.combinations(data.iterrows(),2):
#print("row 1",row1[1])
#index, value = row1[1].items()
for index, value1 in row1[1].items():
arr1 = value1.split(' ')
# print(value1,"smart",arr1[0],arr1[1],arr1[2])
for index, value2 in row2[1].items():
arr2 = value2.split(' ')
#print("pair",arr1[2],arr2[2])
if(arr1[0] != arr2[0]):
continue
dphi = float(arr1[2]) - float(arr2[2])
#print(dphi)
if dphi > np.pi:
dphi = dphi - 2.0*np.pi
if dphi < -np.pi:
dphi = dphi + 2.0*np.pi
dphi_vals = np.append(dphi_vals,dphi)
Going back to our least squares fitting from earlier in the semester, we can extract the modulation.
nbins = 65
bins = np.linspace(-np.pi, np.pi, nbins)
counts, bins, bars = plt.hist(dphi_vals,bins=bins)
uncertainties = np.sqrt(counts)
bins = bins[:np.size(bins)-1]
num_bins = len(bins) - 1
x = bins
y = counts
def cos_mod(x, amp, v2):
"""1-d gaussian: gaussian(x, amp, cen, wid)"""
return (amp*(1.0 + 2.0*v2*v2 * np.cos(2*x)))
gmodel = Model(cos_mod)
result = gmodel.fit(y, x=x, weights=1.0/uncertainties, amp = 3000, v2=0.15)
print(result.fit_report())
plt.plot(x, y, 'o')
plt.plot(x, result.init_fit, '--', label='initial fit')
plt.plot(x, result.best_fit, '-', label='best fit')
plt.legend()
plt.show()
The \(v_2\) value we extract is consistent with the input value of \(v_2 = 0.1\). This is good and reasonably convincing that we don’t have bugs in the code itself.
Now, let’s look at a more interesting scenario:
100 events
50 particles in each event
every particle in a specific event having exactly the same \(v_2\) value but the \(v_2\) value of each event drawn from a Gaussian centered at 0.25 with a \(\sigma\) of 0.10
all other \(v_n\) values are zero
wget_data('https://courses.physics.illinois.edu/phys398dap/fa2023/data/vn_sigma10.txt')
data = pd.read_csv('tmp_data/vn_sigma10.txt', header=None)
print(data)
import itertools
dphi_vals_sigma01 = np.array([])
for row1, row2 in itertools.combinations(data.iterrows(),2):
#print("row 1",row1[1])
#index, value = row1[1].items()
for index, value1 in row1[1].items():
arr1 = value1.split(' ')
# print(value1,"smart",arr1[0],arr1[1],arr1[2])
for index, value2 in row2[1].items():
arr2 = value2.split(' ')
#print("pair",arr1[2],arr2[2])
if(arr1[0] != arr2[0]):
continue
dphi = float(arr1[2]) - float(arr2[2])
#print(dphi)
if dphi > np.pi:
dphi = dphi - 2.0*np.pi
if dphi < -np.pi:
dphi = dphi + 2.0*np.pi
dphi_vals_sigma01 = np.append(dphi_vals_sigma01,dphi)
nbins = 65
bins = np.linspace(-np.pi, np.pi, nbins)
counts, bins, bars = plt.hist(dphi_vals_sigma01,bins=bins)
uncertainties = np.sqrt(counts)
bins = bins[:np.size(bins)-1]
num_bins = len(bins) - 1
#cos_hist, edges = np.histogram(dphi_vals, bins=bins)
#print(edges)
x = bins
y = counts
gmodel = Model(cos_mod)
result = gmodel.fit(y, x=x, weights=1.0/uncertainties, amp = 4000, v2=0.15)
print(result.fit_report())
plt.plot(x, y, 'o')
plt.plot(x, result.init_fit, '--', label='initial fit')
plt.plot(x, result.best_fit, '-', label='best fit')
plt.legend()
plt.show()
#sns.histplot(data=dphi_vals,bins=bins)
Here we get a higher value of \(v_2\). We are essentially measuring the RMS of \(v_2\), \(\sqrt{\langle v_2^2 \rangle}\). When all the \(v_2\) values were the same, that quantity is the same as \(v_2\), but when we add the fluctuating \(v_2\) values, then the RMS value is larger than the average (for a Gaussian the second moment, \(\langle v_2^2 \rangle = \mu^2 + \sigma^2\)).
The warning here is to keep track of what you are actually measuring.
import timeit
from itertools import combinations
foo = [i for i in range(0, 1000)]
def loop_test():
combos = []
for i in range(len(foo)):
for j in range(i + 1, len(foo)):
combos.append((foo[i], foo[j]))
def iter_test():
combos = []
for c in combinations(foo, 2):
combos.append(c)
if __name__ == '__main__':
print(timeit.timeit('loop_test()', setup='from __main__ import loop_test', number=1000))
print(timeit.timeit('iter_test()', setup='from __main__ import iter_test', number=1000))
Fourier Transform
Here we’re going to go over the discrete Fourier Transform. It is defined as: $\(X_k = \sum_{n=0}^{N-1}x_n e^{-i2\pi k n/N}\)\( \)X_k\( is the discrete Fourier Transform, \)N\( is the number of samples, \)n\( is the current sample, \)k\( is the current frequency, \)x_n\( is the sine value of sample \)n$.
\(X_k\) is a complex number and contains an amplitude and a phase (Euler notation of complex numbers).
The inverse transform is: $\(x_n = 1/N \sum_{n=0}^{N-1}X_K e^{i2\pi k n/N}\)$
The problem with this is that it is very inefficient computationally, \(O(N^2)\). If you have many samples, it can take a long time. That’s where the Fast Fourier Transform comes in. This is something that was invented in 1965 by JW Cooley and John Tukey in their paper. What these does is decrease the complexity by exploiting symmetry (this can also be traced back to some unpublished work by Gauss from 1805).
How does it work? Here is a very nice explanation from Jake VanderPlas. Basically the FFT works by exploiting symmetry in the problem.
Starting with the discrete Fourier Transform: $\(X_k = \sum_{n=0}^{N-1}x_n e^{-i2\pi k n/N}\)\( what is the value of \)X_{k+N}\(? That is: \)\(X_{k+N} = \sum_{n=0}^{N-1}x_n e^{-i2\pi (k+N) n/N}\)\( \)\( = \sum_{n=0}^{N-1}x_n e^{-i2\pi n N/N} e^{-i2\pi k n/N}\)\( \)\( = \sum_{n=0}^{N-1}x_n e^{-i2\pi k n/N}\)$
So we can write the discrete Fourier Transform as: $\(X_k = \sum_{m=0}^{N/2-1}x_n e^{-i2\pi k 2m/N} + \sum_{m=0}^{N/2-1}x_n e^{-i2\pi k (2m+1)/N}\)\( now the range of \)n\( is \)M = N/2\( and the range of \)k\( is still \)N\( so now the \)X_{k+M}\( relation above is now relevant. This can be done recursively as far as it makes sense (at a certain point the overhead is no longer with the simplicity). Asymptotically, this is now \)O(NlnN)$.
We’re going to use the numpy FFT and inverse FFT code but the algorithm itself is very simple to implement yourself if you want to make sure you understand it (for real applications, please use a checked and optimized one–they are faster and have handling to deal with edge cases that you might not anticipate).
import timeit
# sampling rate in Hertz
sample_rate = 100
# sampling interval in seconds
sample_interval = 1.0/sample_rate
times = np.arange(0,1,sample_interval)
#the frequencies are integers and the amplitudes can be numbers
freq = 2.
amp = 2.0
x = amp*np.sin(2*np.pi*freq*times)
freq = 4
amp = 2.3
x += amp*np.sin(2*np.pi*freq*times)
freq = 7
amp = 3.4
x += amp* np.sin(2*np.pi*freq*times)
plt.figure()
plt.plot(times, x, 'r')
plt.ylabel('signal')
plt.xlabel('time (s)')
plt.show()
def DFT(x):
N = len(x)
n = np.arange(N)
k = n.reshape((N,1))
e = np.exp(-2j *np.pi * k * n / N)
X = np.dot(e,x)
return X
X = DFT(x)
print(np.shape(X))
N = len(X)
n = np.arange(N)
T = N/sr
freq = n/T
plt.figure(figsize = (5,4))
plt.stem(freq, abs(X))
plt.xlabel("f (Hz)")
plt.ylabel("DFT amp.")
plt.show()
The DFT is symmetric around half the sampling rate or the Nyquist frequency (and the sampling rate here was 100 Hz). Since all the information is contained in the lower frequency part of the spectrum a real implementation of this algorithm will only keep those.
The other side of this is that the signal needs to be sampled at at least twice the frequency that you are interested in. Qualitatively this makes sense
sr = sample_rate
X = fft(x)
N = len(X)
n = np.arange(N)
T = N/sr
freq = n/T
plt.figure(figsize = (12, 6))
plt.subplot(121)
plt.stem(freq, np.abs(X), 'b', \
markerfmt=" ", basefmt="-b")
plt.xlabel('Freq (Hz)')
plt.ylabel('FFT amp')
plt.xlim(0, 10)
plt.subplot(122)
plt.plot(times, ifft(X), 'r')
plt.xlabel('Time (s)')
plt.ylabel('Amplitude')
plt.tight_layout()
plt.show()
And we’d like to verify that the FFT is actually faster than the simple code written above.
%timeit Xfast = fft(x)
%timeit Xsimple = DFT(x)
12.8 µs ± 2.46 µs per loop (mean ± std. dev. of 7 runs, 100000 loops each)
1.13 ms ± 457 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
OK, that’s nice but what about something a little more interesting. What if you have a lot of noise obscuring your data. For our purposes, we’ll put it at high frequency but it could be where ever.
# sampling rate in Hertz
sample_rate = 1000
# sampling interval in seconds
sample_interval = 1.0/sample_rate
times = np.arange(0,1,sample_interval)
#the frequencies are integers and the amplitudes can be numbers
freq = 2.
amp = 2.0
x = amp*np.sin(2*np.pi*freq*times)
freq = 4.0
amp = 2.3
x += amp*np.sin(2*np.pi*freq*times)
freq = 70
amp = 3.4
x += amp* np.sin(2*np.pi*freq*times)
freq = 71
amp = 3.4
x += amp* np.sin(2*np.pi*freq*times)
freq = 65
amp = 3.4
x += amp* np.sin(2*np.pi*freq*times)
plt.figure()
plt.plot(times, x, 'r')
plt.ylabel('signal')
plt.xlabel('time (s)')
plt.show()
sr = sample_rate
X = fft(x)
N = len(X)
n = np.arange(N)
T = N/sr
freq = n/T
plt.figure(figsize = (12, 6))
plt.subplot(121)
plt.stem(freq, np.abs(X), 'b', \
markerfmt=" ", basefmt="-b")
plt.xlabel('Freq (Hz)')
plt.ylabel('FFT amp')
plt.xlim(0, 100)
plt.subplot(122)
plt.plot(times, ifft(X), 'r')
plt.xlabel('Time (s)')
plt.ylabel('Amplitude')
plt.tight_layout()
plt.show()
Now we’ll just go through and zero out the high frequency part of the distribution as a filter. Then we can take the inverse Fourier transform and get back our, now cleaner, signal.
#let's say you want to filter the high frequency part of this
X[50:-1] = 0
plt.figure(figsize = (12, 6))
plt.subplot(121)
plt.stem(freq, np.abs(X), 'b', \
markerfmt=" ", basefmt="-b")
plt.xlabel('Freq (Hz)')
plt.ylabel('FFT amp')
plt.xlim(0, 100)
plt.subplot(122)
plt.plot(times, ifft(X), 'r')
plt.xlabel('Time (s)')
plt.ylabel('Amplitude')
plt.tight_layout()
plt.show()